-
Posts
135 -
Joined
-
Last visited
-
Days Won
2
Content Type
Profiles
Forums
Downloads
Gallery
Posts posted by Götz Becker
-
-
Looks like my problem is similar to this:
http://digital.ni.com/public.nsf/websearch...6256F4D00189B26
Not a "solution" I am happy with, but at least some info on the topic.
-
-
Thx for all your replies.
Next time I have the option, I
-
I am currently learning LV and therefore I try to understand every VI I can get my hands on. After reading some code style guidelines, I try to focus more on style. With this in mind I look at all the sample code I downloaded from all over the web.
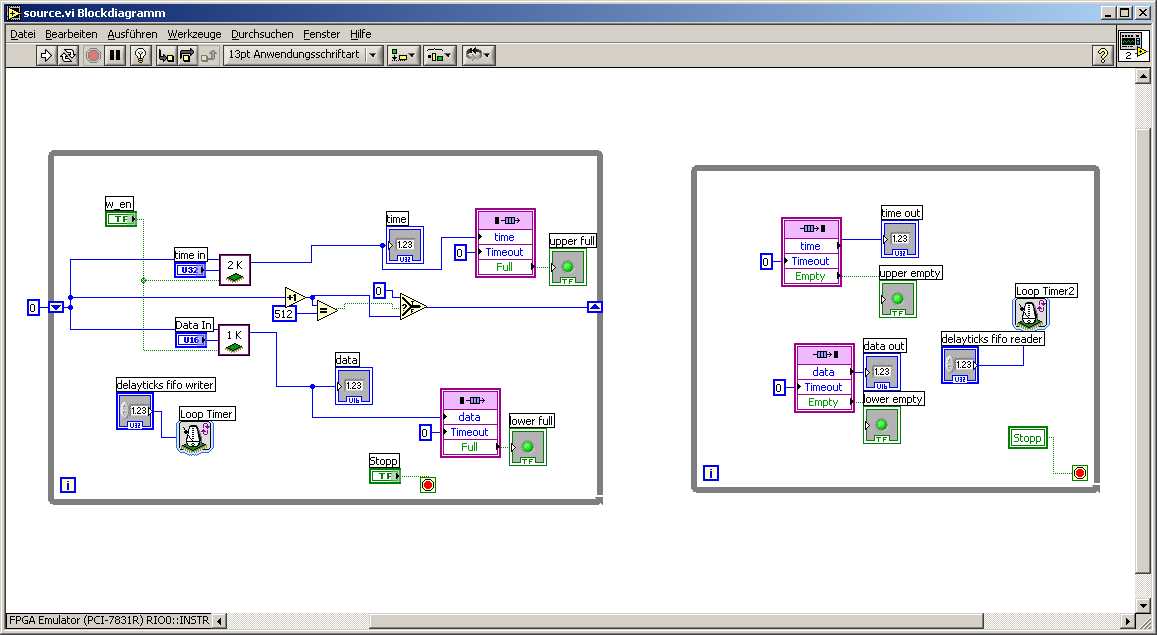
The attached picture shows an example of how weird some VI sometimes look if opened in the german version of LV. Probably this VI looks much better in the english version (e.g. no overlaped subvis).
How badly can this affect portability and reuseability of LV code between different languages versions of LV? Any experiences?
Or is it minor enough to forget about it?
Greets
GB
-
-
Yes I tried with full and sparse data, every time the same.
Don
-
-
Hi all,
i am relativly new to LabView. I am working on an editor for digital data. To display my data i use a Digital Waveform Graph. To do the editing i am using the cursorpos i read out of the property node. But when I try to set the cursorpos either by typing in a numerical value in the cursorpalette or programatically by writing to the property node, I end up with wrong values!?!
For example, the graph diplays data from 0 to 10000, I type 487 for X-cursorpos and LabView alters my input to 480 and places the cursor at 480.
I tried some values
[typed-input -> altered to]
487 -> 480
488 -> 495
500 -> 495
502 -> 495
503 -> 510
518 -> 525
Is this the normal behaviour?
Greetings
Goetz Becker



Strange behaviour using queue
in Application Design & Architecture
Posted
Thx for that information!
I already gave up hope to ever fix a bug in my current app. Good thing I already had all my queue accesses wrapped in subvis .
.
But I have seen this bug in my standalone version too!
Greetings
Goetz