-
Posts
317 -
Joined
-
Last visited
-
Days Won
7
Content Type
Profiles
Forums
Downloads
Gallery
Everything posted by gb119
-
So the Jupyter client protocol is probably not the way to pass large chunks of data between Python and LabVIEW - it's intrinsically a text based messaging system (since it's really designed for interacting at a console like device with the Python kernel). If you Python kernel is running on the same machine as the LabVIEW code, then probably the simplest way to transfer the data back and forth is to write it to a file. Since even a fast SSD is not particularly fast, you might want to setup a RAM disk for this purpose. You would then use something like this JuPyter client to import some module of your code and then call some function, passing the name of the input file. You make that function return the name of the output file and your LabVIEW code reads it back in. On unix-like systems you might be able to get away with a named pipe for the transfer mechanism (I've no idea if one can do that on Windows or not...). If the Python Kernel is remote to the LabVIEW then clearly you're going to need send it over the network. It's quite easy to write a Python TP/IP server, so I guess you could trigger a function with the JuPyter client that will launch a server. You'd need to manage the process of serializing the data, but if you don't use the JuPyter messaging protocol you can at least send it in a fairly compact binary format. I sort have an idea that it should be possible to use something like the Msgpack binary serializers to package LabVIEW data types and transfer them and write a deserializer in Python at the other end that unpacks them - have the feeling that the numpy in-memory sructure and LabVIEW's flattened double/single arrays might actually be compatible which would make it much simpler. The JuPyter message protocol sort of has the mechanisms to do this builtin via the concept of a Comm - it just needs time to work on it...
-
The pother advantage of the Map type over the variant attributes is that it serialises correctly with the Variant to XML string node. Very annoyingly, in LV2019 it's not supported by the NI Variant to JSON node (I don't know if that'#s been corrected in LV2020 - I'm a little nervous about mixing the Community edition for my personal play-time projects with my academic site licensed LV installs for work and playing...).
-
For the first of those, you'd want the LV2018 branch. Jim K recently brpught some infelicities in the code to my attention - which of cousre becomes an incentive to fix things (thus proving Rolf's point!). I'm still having an issue with files> 2Gb - there's something I don't understand when dealing with 64bit files (on 64bit Windows and 64bit Labview). Not helped by the fact that testing is slooooooow.
-
Sorery dunno what has happened there - I created the snippets in LV 2019 64bit, saved as aPNG files and dragged it ontot he post in Chrome - perhaps somewhere between Chrome, and LAVAG it stripped the embedded LabVIEW code? From your comments, I think the answer is simply that variant attributes are in fact quite fast and at least for string keys is the way to go for now.
-
So for my usual use cases I do want the keys to be strings - the type cast is just a means to generate a random string. Its also the same in both versions of the benchmark so cannot on its own explain why the map is slower. A better benchmark would probably be to pre-calculate the keys and then use the same keys for both map and variant attribute.
-
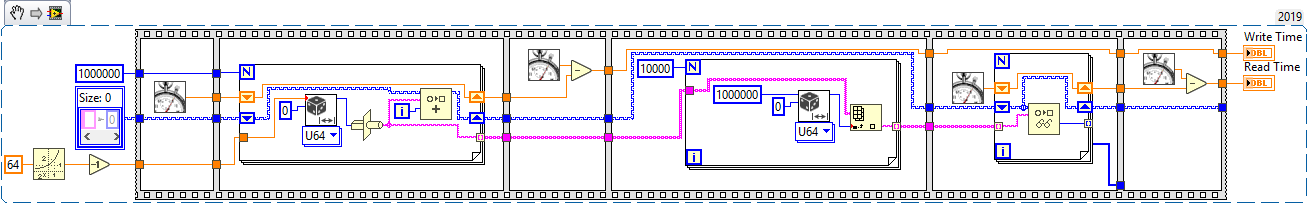
So I got very excited when I saw that LabVIEW 2019 has a new native map type (aka hash array, associative array, dictionary) and so decided to have a play and see how it compared to my home-rolled LVOOP has array that uses variant attributes and I must admit that I'm slightly underwhelmed.... I've now benchmarked the 2019 native map class and a simple variant attribute by creating maps of 1 million elements of randomly generated 8 byte keys and then reading back 10,000 randomly selected elements and fairly consistently the native map is about an extra 50% slower than the variant attribute for both read and write operations. I'll admit my benchmark code is quite naive, but it's still a little disappointing that there is quite this performance hit.... Can any of the NI folks here comment on the performance - is it just that in fact variant attributes are blazingly fast? I know I shouldn't be churlish and it's really great that 2019 has so many new features and a native map and set type have been long overdue... Edit: and yes I've spotted that the variant type conversion was wired wrong and I should have been generating an array of 10000 I32 not error clusters - but no it doesn't make a significant difference....
-
But surely all of these are slower and more memory wasteful than transposing which simply twiddles the order and offsets of the pointers that are used to index the block of memory in which the array resides?
-
So what you want to do is not so straightforward. The thing is that the Jupyer Client code is not interacting with the notebook directly. When you start the Jupyter notebook you are both starting the frontend that runs in a web-browser, but also starting a kernel process in the background that the front end interacts with. What my code does is provide another front end that can talk to the same kernel backend process - so both front ends can change the state of the kernel (i.e. create variables etc) and interrogate the kernel about its state - so in that way the two frontends are aware that 'somebody else' is messing with 'their' kernel. But, the two frontends don't talk to each other - so LabVIEW cannot directly manipulate what the notebook displays. The only exception that I've found with a little minimal playing is that if you invoke the %matplotlib notebook magic in the notebook and then create a new blank figure in the notebook, then you can plot into tat figure from the LabVIEW end i.e. In the notebook I do: from matplotlib.pylab import * x=linspace(-pi,pi,361) y=sin(x) %matplotlib notebook figure() and in the LabVIEW client I do: plot(x,y,'b+') Then I get a plot of the sin function in the notebook - but I think that is more or less an accidental consequence of how the matplotlib notebook backend works. I started wondering what you need is a LabVIEW JuPyter kernel - ie. something that could interact with a a LabVIEW process via a notebook - that would be a fun project, but just not this one. Thinking further, however, I think your use case is actually mix of both kernel and client - use case 3) is pretty much a LabVIEW Jupyter kernel, use case 2) seems to require that the notebook also has access to a Python kernel and is more like having a Python module that knows how to speak to a LabVIEW kernel. Use case 1) is a bit of a mixture.... What this code is best suited for is off-loading processing of LabVIEW data into Python routines, or allowing a LabVIEW front end to interact with a Python based system - e.g. some sort of distributed control system with a Python kernel sitting in it.
-
So this is where it gets trickier. Intrinsically the kernel-client protocol is geared around sending strings to the kernel and by and large getting strings back. This makes sense if one thinks the client is essentially a terminal with a keyboard, a screen and a human. So trivially you can express the data to be sent to Python from LV as an assignment statement "x=3.141592654" and have that executed on the kernel and it will create variables in the kernel's namespace - but it's hardly efficient if what you want to do is send a moderate sized 2D array of floating point numbers over. I think the solution is to implement a pair of Comm[1] objects to send and receive custom messages[2] in which data can be encoded in a more efficient way and used to manipulate the globals() on the Python side and request specific variables to be sent back to Python. I've had a look around at various 'binary json' serialises and liked the look of ubjson[3] the most (ideally I'd implement the Python pickle algorithm in LabVIEW, but that didn't look fun!). That would solve the how to encode the data for transfer part of the problem. The actual mechanics I think will involve a CommunicationsChannell LabVIEW class that has methods to squirt the python needed to create the Comm objects at the kernel end, deal with the kernel requesting to open it's side of the communications back to the LabVIEW and then methods to send arbitary LabVIEW data and request Python data and map it back to LabVIEW types. Finally it will need to implement comm-open, comm-close, and comm message types - but they're basically easy given the class hierarchy that already exists for messages. All of which is great, but I'm running out of Easter vacation, about to hit the summer exams and I'm the departmental exams officer so have negative free time for a few months! [1] https://jupyter-notebook.readthedocs.io/en/stable/comms.html [2] https://jupyter-client.readthedocs.io/en/latest/messaging.html#custom-messages [3] http://ubjson.org/
-
That solves part of the problem, but not all of it. Using the Convert State for Save ability lets you stop writing volatile state information to disk, but it doesn't stop LabVIEW from thinking that the host vi (i.e. the one you've put the XControl into) requires saving. That happens as soon as you toggle the state changed boolean in the action cluster. You need to have an entirely separate means of storing volatile, per instance, state and then not touch the state boolean in the action cluster unless you really do mean to record a change that would require saving. I've been using variant attributes on a variant stored in an LV-2 style global, but I guess one could use 1 element queues, or even, gods forbid, a global variable! That said, I;ve just realised that I ought to have coded the Uninit Ability on my XControls to clean up the per instance variant attributes when the host vi goes out of memory....
-
That may be a while - but the package file itself in the top of the thread.... Edit: Thinking about, because I'm dependent on the ZMQ bindings which are not available on the NI Tools network, I'm not sure I can put this package (and the SHA-256) library on the NI Tools network either - so it will always need to be installed from manually downloaded vipm files.
-
Ok, bit of Easter holiday coding today. Version 1.1.0 should allow connections to remote and already running kernels (well it does for me), and will only issue kernel shutdown messages if it started the kernel itself. To connect to a remote kernel, you can either manually fill in a cluster of port numbers etc, or simply paste the json from the connection file or (if you have an existing front end to the kernel) do: from ipykernel.connect import get_connection_info print(get_connection_info()) If starting kernels on another machine, remember to tell them to bind to an IP address that isn't localhost e.g. ipython kernel --ip=u.x.y.z and make sure the firewall will let the ports through.
-
Hmm, there was a problem I had there but I thought the version I packaged had fixed it. My current development version should find that path - but it depends quite a lot if you have multiple Pythons installed on your machine. BAsically there doesn't seem to be a bullet proof way of getting the correct path in Windows.... That's a sensible idea - it's going into the development code. That's largely a result of the test client being mainly aimed at debugging the protocol and for testing message handling rather before moving on to code to more tightly integrate LabVIEW programs with the remote kernel. That said, I'm in the process of adapting the client to allow different methods of locating and connection to the kernel and that will include suppressing the kernel shutdown message on exit. I'm also (very slowly) working on an implentation of a LabVIEW universal-binary-json serialiser/deserialiser with a view to creating some custom ipython messages for transferring binary data efficiently between LabVIEW and Python. The idea is that the LabVIEW client would create message handlers at the Python end that would allow LabVIEW data to be pushed directly into the Python namespace or to request python data to be sent back to LabVIEW. Don't hold your breath though, the day job comes first...
-
Is this the problem of handing 'volatile' state where you need to track state during the run-lifetime of the XControl but do not want to persist it to disk? Using the display state and setting the corresponding boolean in the action cluster then marks the host's dirty bit....? For my XControls I ended up storing a copy of the state in an LV-2 style global. To support multiple instances of the XControl, the LV-2 global is actually a variant and I store the state in attributes of the variant using the container refnum (cast as a string) to provide the attribute name. The first thing the facade does is to read the state and the last thing is write it back to the LV-2 global. XControls are a bit of a pig, on that I guess everyone can agree....
-
You ate more likely to get help if you can describe' (or better still post) what you have tried to do so far. Also, your question is not well defined - are you supposed to interface with some hardware to detect the traffic. What is the desired response to detecting traffic -- signal traffic to go or to ensure tje traffic is stopped? Is this supposed to be controlling an intersection - if so how many ways? Do you have pedesyrian or cycle lanes? Does some traffic get priority? What is the maximum waiting time at a stop light?
-
The original article has been updated with a new version of the JuPyter Client build as a VI Package. This version has switched the parser to JSONText and removed the OpenG dependencies, and fixed up a bunch of bugs. The example client application gets installed into the example finder. It's still far from production ready code...
-
That was the direction I was thinking in - when I've finished unbreaking the effects of changing JSON parser 🙂
-
So for transferring largish chunks of floating point data (e.g. image data or similar) JSON is quite unwieldy (and I worry about losing precision when round tripping to/from ascii data). I haven't looked at it in enough detail, but it seemed to me that there was a good chance that the numpy representation of a double float and LabVIEW's might be sufficiently close that one could do a fairly efficient pack/unpack operation. I take the point about the speed of the JSON serialisers - the choice was more influenced at this point by what I was used to workign with than speed. I'm looking at replacing it with yours and ijn the process keeping more of the message as raw JSON strings rather than storing in arbitary lusters in variants....
-
For a while I've been tinkering with the idea of building a LabVIEW client that could to talk to Jupyter kernels for interfacing with Python having been previously a user of RolfK's OpenG LabPython package. Although this, and now the native LabVIEW 2018 Python support have many uses (and indeed I use them in my 'production' code), there were a few things that a Jupyter kernel client can do: Not be tied to particular versions of Python - LabPython got stuck for me around 2.7.10 and I think was fussy about which compiler have been used. The 2018 native support is restricted to 2.7 or 3.6 I believe (3.7 defintiely doesn't work) Not being tied to the same 32/64 bits of LabVIEW Being able to offload the Python to a remote server, or go cross platform I haven't investigated the Enthought package (too much hassle to get a new vendor set up on my University's purchasing system and not really able to justify spending tax payer's money on playing!) which I suspect might be doing something similar. Anyway, the attached zip file is a proof of concept - it includes a test vi that will try to find an ipython executable and fire it up and you can then interact with it. There's lots of things not properly tested and probably a slew of bugs as well. To run it you need several dependencies: OpenG Toolkit libraries, particularly the LabVIEW Data, string, error and array libraries The JKI JSON library - I had to pick a JSON serialiser and the JKI one seemed as good as any and better than some... The JSONText JSON serialiser library available via VIPM The Zero-MQ Labview bindings - libzmq is the underlying network transport used in Jupyter and there is an excellent LabVIEW bindings library for it. The attached SHA256 implementation so that the communications messages are properly HMAC signed. LabVIEW 2018 - sorry I'm only writing in 2018 now and this code uses malleable vi's with type specialization and asserts in use - so it may not be easy to backport There's a few things that I'd still like to figure out - primarily the client protocol is very much focussed (reasonably enough) around the idea that the client is sending strings and is interested in string representations of data.I'd like to figure out an efficient way to transfer largish LabVIEW data structures backwards and forwards. I think this probably means developing a custom message handler and registering it with the kernel when the code starts and writing some Python 'flatten to string' and 'unflatten from string' code - but that's only this week's concept.... If you use it, please note that this probably only alpha quality at best - it may or may not work for you, it may not be safe to use, If it causes any loss or damage or eats your cat then it's not my fault.... Edit 6th March 2019: I've switched the JSON parser to JSONText, found and fixed a few bugs, managed to build a VI package for it that should have the correct dependencies and installs the example client in the LabVIEW example finder. Edit 19th April 2019: Added more options to connect the example client to remote and already running kernels (and not to shut them down on exit!). Some other fixes as well. Edit 11th Arpil 2020: Updated the SHA256 version to one that can correctly hash files without reading the whole thing into memory. university_of_leeds_lib_jupyter_client-1.1.0.6.vip university_of_leeds_lib_sha256-1.1.2.7.vip
-

Estimate magnitude/phase on single frequency
gb119 replied to Axelwlt's topic in Object-Oriented Programming
I would use a lockin amplifier with some sort of comms interface:-) Failing that you could simply implement the same algorithm - multiply your signal by sine and cosine waveforms at the refetence frequency, low pass filter, calculate the mean and feed into x,y to polar co-ordinate convertor. Altetnatively, up-sample the signal, feed into an FFT, index out the right (complex) element and covert to r,theta. -
Having to use the accessor vi rather than a property node tripped me up - I can see how the compilation issues mean not accessing the private data directly, but not the difference between calling an accessor vi directly or inside the property node.....
-
I've seen variants of 'Vi Failed to Compile' with projects involving complicated class hierarchies and class instances as data members of other classes etc. Nothing particularly new - I've seen this in versions of LabVIEW from 8.6 onwards - usually simply opening the offending Vi 'standalone' is enough to get it to compile then everything is ok until the next time that LabVIEW decides it needs to compile all my code. Now in 2018 64bit I've got a slightly odd one where when I open my top level Vi, LabVIEW decides that the Actor Framework isn't compiling, but if you open that first and then load the top level Vi everything is fine... I find it incredibly hard to debug the issuse as it only happens when one has a sufficiently large project. Keeping compiled code separate seems to make it more likely, as does having very large class hierarchies or classes with private data containing classes. Actor Framework also seems to be a contributing factor. The error message is almost as helpful as the old MacOS installer error "There was a problem with the hard disc, please use a different one....."
-
Assuming that "crap" is not a vialbe option, I like "brittle" too. "fragile" to me would imply code that is susceptible to runtime errors as opposed to one that is easily broken by subtle changes in editing. Perhaps "chaotic"in the maths sense might suit?
-

LabPython 1050 error shown when import sklearn library
gb119 replied to damagor's topic in OpenG General Discussions
Well, the screenshot you showed implies that Python threw a Syntax Error exception - although I can't immediately see why. There are a couple of issues that I can see in your code though: 1) You've got several Windows style paths quoted in strings there, but \ in Python is an escape character - to include \ in a string you need to either do "C:\\Python27\\Lib" or else use a raw string r"C:\Python27\Lib" 2) There is a major limitation to do with thread safety between Python and LabVIEW - LabVIEW is intrinsically multi-threaded and some Python libraries - such as numpy - are not. This can lead to seemingly random crashes in LabVIEW - so not your current problem, but likely to become one... There's been quite a bit of discussion in various threads here on this problem by folk who know more than me about the internals of both LabVIEW and Python.... (and I've just spotted, does np.loadtxt close files when passed an open filehandle - if not then you'll be leaving files left open by your script.) -

OpenG Package: LabPython v4.0.0.4 Install Error
gb119 replied to damagor's topic in OpenG General Discussions
As well as having 32 bit LabVIEW, is your Python 32bit as well? What distribution of python are you using (or vanilla python 2.7?) I've failed to get recent versions of anaconda Python to play nicely with LabPython whereas vanilla 2.7.10 from python.org was ok. Sometimes I find that running VI Package Manager via 'Run as Administrator...' is necessary even though my user account is a member of the Administrators Group. The error looks like it's a file permissions problem, so probably the latter is the first place to try...