-
Posts
264 -
Joined
-
Last visited
-
Days Won
26
Content Type
Profiles
Forums
Downloads
Gallery
Everything posted by GregSands
-
Not sure what this has to do with LabVIEW - perhaps you're better to ask on the Grafana website.
-
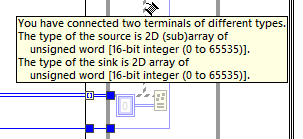
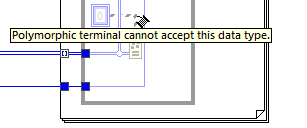
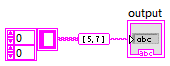
I've started trying to use .vim files a little, and have a couple of questions: 1. Looks like a typedef doesn't match with the same structure without a typedef, correct? I would have thought that if the internal data matches, it should be ok. 2. I get this interesting mismatch between an array and a sub-array: But if the connection is the other way around, I get this error: I would have hoped that either (or both) would be ok. I'm not in the beta, so don't know if this has already changed, or is likely to?
-
Sounds reasonable. Even NI only supports four versions for some of its modules (e.g. Vision Development Module), so I've finally jumped from 2012 to 2017. Is there a way to keep the current install available in VIPM if using LabVIEW <2013?
-
My first thought - I wonder whether one of the 3D Graph (I find the old NI 3D Graphs a little easier than the rewrite) or 3D Picture controls might work - unfortunately there seems like there might be a fair bit more capability hidden out of sight than can be easily accessed. At the very least you might be able to appear like a bar has two different colours by an appropriate 3D Bar Plot, even if you don't get the gradient in between. 3D Picture controls are based on OpenGL, so theoretically it should be easy to do what you want there, but I've never had much success doing anything complex with 3D Pictures.
-
Seeing a Malleable VI (.vim) implementation
GregSands replied to GregSands's topic in LabVIEW General
Yes, that's what I've been doing, and it's ok, though suboptimal as you say. I'm looking forward to seeing the development of Malleable VIs - I've held off on trying to convert some of my XNodes to VIMs, but if things stay fairly stable, I'll have to give it a go. -
We've lost something useful in the "official release" of Malleable VIs (.vim files aka VI Macros) in LabVIEW 2017. In previous versions, because VIMs were built around XNodes, then you could right-click the XNodeWizardMenu to look at the Generated Code given a particular wiring. There's no such option in 2017, even with the appropriate LabVIEW.ini keys. Is there another ini key that provides a similar functionality again? I find it a useful check that the VIM is coded correctly. The closest is to "Convert Instance VI to Standard VI", however that removes the VIM.
-

How to draw circles and lines in Intensity Graph?
GregSands replied to wio_letta's topic in LabVIEW General
Probably easiest to use the PlotImages.Front property, which allows you to create a picture with all the lines and text, and overlay it on the Intensity Graph. Also, if you look in the Classic Graph controls there is a Polar Plot control which contains some subVIs for drawing the polar diagram that might be a good starting point. -
One approach that is similar to, but a little more robust than, your original method is to use the Peak Detection vi which fits a quadratic to the data, and returns a fractional index. Here I've used it just to shift each waveform so that the peak is at zero, but you could use the shift information in different ways. Here's a noisy signal, and increasing the width of the fit seems to cope with it ok.
-
Tim's right in that you need to provide a lot more information - the appropriate filters and parameters are very dependent on the raw data. Having said that, for the purpose you describe (which I'm guessing is signal visualization rather than signal analysis) the first filter I will tend to try is the Savitzky-Golay filter, though again the parameters can be quite sensitive. It's essentially a piece-wise polynomial fit. There is also a Pt-by-Pt version which is useful for real-time data processing as it is acquired.
-

Area within a curve - Green / Kelvin-Stokes Theorem
GregSands replied to gyc's topic in LabVIEW General
In fact there's Polygon Area.vi in the Mathematics/Geometry/Computational Geometry palette. Slightly different but equivalent implementation. Typing "area" into Quick Drop found it immediately -
No I've not used netCDF. I presume that if netCDF uses HDF5, then it is just a particular file layout (combination of dataspaces and attributes) that ensure that all the required information is stored in a well-defined place. I agree that using/supporting HDF5 would be of more use to more people. The h5labview code is here (<500 lines) if you wanted to see what few things it is doing. It's all open-source, so I'm sure Martijn would be appreciative of any extra development. From the FAQ :
-
I've used HDF5 in two situations (so far) for quickly capturing large 3D imaging datasets (XYZ or XYt) - I've used the open source h5labview package which has worked very well for me. The other LabVIEW package (not open source) that wraps HDF5 is Live HDF5, but unfortunately the two do not play nicely together, so it's not possible to install both at the same time to evaluate. The HDF5 libraries are mostly wrapped directly, with a small interface dll just for handling some of the memory allocation issues. HDF5 is similar to TDMS, but much more flexible, and not just in array dimension - H stands for Hierarchical, which basically means anything can go anywhere - data, attributes (metadata), groups, ... - even seamlessly across multiple files. What I think is most flexible about it is that you can have different arrangements of your data in memory and on disk, and HDF5 handles the mapping between. Needless to say that can also get very complicated, so I tend to stay with fairly simple mappings! Used simply, it's straight-forward, but there's a lot of power underneath. I've said more than once that I wish NI had built their data format on top of HDF5 rather than developing their own proprietary format - in the way that Matlab now uses HDF5 as the internal format for .mat files.
-

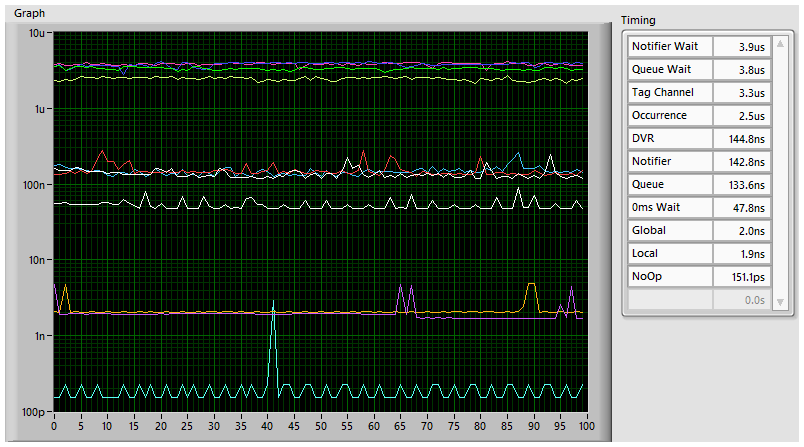
Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
So there seem to me to be three questions related to this thread: Are Tag Channels appropriate to use in high-throughput loops? If not, that is a fairly significant reduction in the use cases for Tag Channels (i.e. I need to work out in any given situation what the loop time is in relation to an occurrence check). But if Tag Channels should be considered for fast loops, is the check of an occurrence the appropriate way to implement the Tag Channel? Could it use Get Notifier Status instead? If checking an occurrence is best, is the OnOccurrence() function able to be made faster? Because at the moment, it does look as though it is not feasible to use a Tag Channel where speed is critical. I'm assuming that other Channel implementations (e.g. Streams) don't have this issue - it looks as though they only check an occurrence if they need to wait because the stream is full. -
You could also do all of this using just the Vision functions, without using arrays or overlays, and it's probably much easier. Have a look at the IMAQ ReplaceColorPlane function, which allows you to place grayscale images in separate planes of a color image. To translate the images, look at the IMAQ ImageToImage function which is used to place a small image at a specified location within a larger image.
-

Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
Thanks. This means I guess that the value shown above will be the best-case (for a single reader) and is comparable with a local variable read. -

Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
I didn't know that, interesting. I modified the test code to check all these options, with these results (median time per iteration): -

Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
I was thinking "this can't be right, maybe that's only on your sbRIO", but I checked on a Windows desktop (just creating a new case in your timing test above), and checking an occurrence takes ~75% of the time of checking the Tag Channel. I've almost never used occurrences, but I note in the Help that "National Instruments encourages you to use the Notifier Operations functions in place of occurrences for most operations." Perhaps Channels should follow this advice! Unless of course there is some reason that an occurrence is required. -

Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
OK, that all looks interesting, even reassuring, but does it explain why the Tag Channel appears to be so slow? And is it specific to a Tag Channel, or the same slowdown for any type of Channel? Given it's a new technology it's not too surprising that there are some optimizations yet to come, but the degree to which it is slower is concerning. -

Boolean Tag Channel vs Notifier Performance
GregSands replied to infinitenothing's topic in LabVIEW General
Opening up the Tag VIs, it doesn't look as though there's a call by reference (except when it is Instantiated) - it's a straight call inside the channel code. But there is a Lookup Channel Probe on every call - I wonder if that's what's slow? Nothing else looks particularly unusual or time-consuming. -
There weren't many dependencies, so here's a new version without any. In some ways it's cleaner CompactIndexArray_1.1.zip If anyone wants to modify or extend it, you're very welcome.
-
Oops, sorry, I should have mentioned that dependency! Thanks Mikael, and Gavin for the tools. Couple of OpenG VIs used also I see.
-
In response to this Idea, and to refresh my xnode coding, here's a compact form of Index Array with fixed indices (double-click to change). Thanks to hoovah for his XNode Editor! CompactIndexArray.zip
-
Installing HDF5 on Windows is a breeze, no problems there. And there is plenty of documentation and examples online, but very little describing using it in LabVIEW. The question is whether its worth dealing with the learning curve which, in the end gets you somewhere much more powerful, but at the cost of time and complexity. I should also have mentioned that h5labview is a wrapper around the HDF libraries, and only includes a subset of all that it can do - a pretty reasonable subset to be sure, and the developer seems quite open to ideas. I think the other LabVIEW option (Live HDF5) has similar restrictions in capability. One annoying thing is that both wrappers cannot be installed simultaneously due to filename conflicts, so it's hard to directly compare them. And yes, HDF5 attributes are similar to TDMS properties, but more so - they can be arbitrary data themselves, not necessarily just strings. So it's easy to store, for example, a settings cluster as an attribute to its associated data.
-
I also use TDMS at times, and it has some other advantages, such as being able to write directly from DMAs (say from an FPGA) or DVRs, or configuring to write internally using DAQmx logging. Where the data is written and read only in LabVIEW or Diadem, it's a natural option, but it's not so suitable if other non-NI software is used for the data analysis - the connection to Excel and Matlab is limited at best, and for anything else you seem to be on your own, or needing to interface through another file format. Another option which is more standardized is HDF5 - I'm a fan of the h5labview library, but there are other alternatives as well. HDF is extremely flexible around the hierarchical relationship between the data and the storage format (so much so that it can get quite convoluted if you want it to), and is at least as fast as TDMS for reading/writing. Some advantages include the ability to read (or write) with arbitrary offsets/steps, even in multi-dimensional arrays, and the ability to add attributes at any level of the storage (file, object, data group, data, etc). Downsides are just getting your head around the options especially with data spaces (a mapping between your data array format and the data file format) - in some ways it's too flexible, although basic use is quite straightforward. Here's an example image from the h5labview website: I use HDF5 very successfully for large 4D image data, because it makes it easy to access just the data I want, but for 1D data streams I would tend towards TDMS as the first option. I must confess to only having used SQLite once for text logging - maybe I need more of a look at it too! What I really wish is that NI had built its data storage formats around HDF5. There are few, if any, features of TDMS that could not be achieved with a file format compatible with HDF5. A similar idea has reached 55 kudos to date, which shows a decent level of support for a largely technical idea. Note that Matlab's .MAT file format is now HDF5.
-

MOV to AVI or intensity array?
GregSands replied to Bill Gilbert's topic in Machine Vision and Imaging
The link I posted does exactly this on Windows (though with ffmpeg) and streams the video through a pipe into LabVIEW as YUV binary data, which is then converted on the fly into RGB images.- 14 replies
-
- conversion
- video
-
(and 2 more)
Tagged with: